 |
CARBOXYPEPTIDASE A
THE ACTIVE SITE
|
In this exercise you will use RasMol to look at the active site of Carboxypeptidase
A with and without a substrate.
The tutorial is divided into two parts. In Part One, you will
isolate the active site of CPA with no substrate present. In Part Two,
you will isolate the active site with the substrate Glycyl-Tyrosine in
the active site.
NB. It may be worth your while to obtain a hard copy of this tutorial
exercise to avoid having to switch back and forth between the Browser and
RasMol windows. Choose 'print' from the file menu if you have access to
a printer. Students working through the exercise in a tutorial session
should have a copy with their tutorial handout.
You will have some questions to answer along the way! These will
be indicated by the Q/A icon  .
When you see this icon,
single click on it to go to the appropriate
questions.
.
When you see this icon,
single click on it to go to the appropriate
questions.
To view answers to the questions, your web browser must be capable of
recognising JavaScript, eg. Netscape 2.0x. (although my experience indicates
that Netscape v2.02 will only work 50% of the time). When you click on
the "Check Answers" button, a separate window should appear with the correct
answers. An alternative
version of the tutorial is available for web browsers not capable of
recognising JavaScript.
For further information on any of the commands used in this tutorial,
consult the online RasMol
Manual

PART ONE - The Active Site
-
Step One - Begin RasMol
-
Begin Rasmol by single clicking on the Rasmol icon (above). Two
windows should appear - the graphics window (with a wireframe image of
Carboxypeptidase A) and a command window with the Rasmol prompt :
-
RasMol >
Organise your windows so that you can see the graphics window and the command
window, try resizing the command window into a short, wide window, then
position it at the bottom of the screen. You will be entering commands
at the Rasmol prompt. Remember - you must select the command window (click
in the window with the mouse) in order to type in the commands.
-
Step Two - Isolate the Active Site
-
You are now going to isolate the active site of carboxypeptidase A. To
accomplish this you must issue a series of commands to define the active
site members and display them in the window. It is often more convenient
to write a text file (known as a 'script' file which contains the
commands you wish to execute. The commands can then be executed all at
once with the RasMol 'script' command. We have written a small script file
for you (as1)
which will define a set known as 'AS' to include members of the CPA active
site. If you are accessing this tutorial from anywhere other than the Biomedical
Sciences Mac Lab, choose the link to the script files on this page and
follow the instructions given.
The following commands should leave only the residues involved in the
active site within the graphics window :
-
script as1
(enter)
-
restrict AS (enter)
-
colour cpk (enter)
-
Colour by atom type (if changed from original default)
-
Step Three - Members of the Active Site
-
Position the active site in the center of the graphics window:
-
translate x -5 (enter)
-
translate y -45 (enter)
-
Now try zooming in on the active site.
-
zoom 300 (enter)
-
Which amino acid residues are involved in the active site?
First, let's pin-point the Zinc atom which is essential for enzymatic
activity :
-
centre zn (enter)
-
select zn (enter)
-
colour cpk (enter)
-
script cpa5zn (enter)
The cpa5zn
script uses the 'monitor' command to identify the atoms which coordinate
zinc, note that a water molecule participates, although only the Oxygen
atom is displayed.
-
Next, identify the residues which coordinate the Zinc atom :
-
select glu72, his69, his196 (enter)
-
Important Note! It is sufficient throughout this tutorial
to enter the numbers of the amino acids without the three letter code.
We will include them here for your information.
-
colour green (enter)
-
script as3
(enter)
-
A script file labels the appropriate residues
-
Finally, identify the amino acid residues which interact with the substrate
:
-
select arg127, asn144, arg145, glu270, tyr248 (enter)
-
colour yellow (enter)
-
script as4
(enter)
-
A script file labels the appropriate residues

Single click on the image if you wish to check your work.
-
Step Four - Experiment
-
Experiment with the RasMol menu in the graphics window. First, select the
active site :
-
select AS (enter)
Then try some of the Display options - eg: Spacefill
and Colour options -eg: CPK
 Test
your knowledge!.
Test
your knowledge!.
-
Step Five - End of Part One
-
Close the current molecule by typing ZAP in the command window.
PART TWO - The Active Site With Substrate (Glycyl-Tyrosine)

-
Step One - Begin RasMol
-
Begin Rasmol by single clicking on the Rasmol icon (above). Two
windows should appear - the graphics window (with a wireframe image of
Carboxypeptidase A) and a command window with the Rasmol prompt :
-
RasMol >
Organise your windows so that you can see the graphics window and the command
window, try resizing the command window into a short, wide window, then
position it at the bottom of the screen. You will be entering commands
at the Rasmol prompt. Remember - you must select the command window (click
in the window with the mouse) in order to type in the commands.
-
Step Two - Isolate the Active Site with Substrate
-
You are now going to isolate the active site with glycyl-tyrosine as the
substrate positioned within the active site.
Once again, we have written a short 'script' file (as2)
which defines a set known as 'AS' to include members of the carboxypeptidase
A active site and the substrate glycyl-tyrosine.
The following commands should leave only the residues involved in the
active site within the graphics window.
Commands:
-
script as2
(enter)
-
restrict AS (enter)
-
Step Three - Members of the Active Site
-
Position the active site in the center of the graphics window.
-
translate x -5 (enter)
-
translate y -45 (enter)
-
Now try zooming in on the active site.
-
zoom 300 (enter)
-
Which amino acid residues are involved in the active site?
First, let's pin-point the Zinc atom which is essential for enzymatic
activity :
-
centre zn (enter)
-
Molecular rotation now centred on the zinc atom.
-
select zn (enter)
-
script cpa3zn
(enter)
The cpa3zn
script uses the 'monitor' command to identify the atoms which coordinate
zinc, note that the substrate displaces the water molecule in the active
site.
-
Next, identify the active site residues which coordinate the Zinc atom
:
-
select glu72, his69, his196 (enter)
-
colour green (enter)
-
script as3
(enter)
-
A script file labels the appropriate residues
-
Finally, identify the amino acid residues which interact with the substrate
:
-
select arg127, asn144, arg145, glu270, tyr248 (enter)
-
colour yellow (enter)
-
script as4
(enter)
-
A script file labels the appropriate residues
-
The remaining residues belong to the substrate.
-
script as5
(enter)
-
A script file labels the appropriate residues
-
Which atom in the substrate becomes coordinated to the zinc atom? You may
want to check this reference.
Use the mouse to click on the zinc atom, then the atom in the substrate
which coordinates the zinc atom (it may be easier to pick atoms if the
substrate is displayed as a Ball and Stick model). Information about each
atom is written into the command window:
-
{atom} {atomno} {Type: residue} {residue no} [Chain: identifier]
-
Notice that the second field contains the atom number. Use the monitor
command to visualise the coordinating bond between the appropriate atoms,
ie.:
-
monitor atomno1 atomno2
Where atomno1 is replaced with the Zn atom number and atomno2 is replaced
with the appropriate atom number from the substrate.

Single click on the image if you wish to check your work.
-
Step Four - Experiment
-
Experiment with the RasMol menu in the graphics window. First, select the
active site :
-
select AS (enter)
Then try some of the Display options - eg: Spacefill
and Colour options -eg: CPK
 Test
your knowledge!.
Test
your knowledge!.
-
Step Five - Active Site Environment
-
You should be aware that carboxypeptidase A hydrolyses carboxy-terminal
amino acid residues of proteins with bulky aliphatic or aromatic amino
acid residues more quickly than it does other carboxy-terminal residues
of proteins. You are now going to look at the environment about the amino
acid side-chain of the substrate (phenolic group in this example of glycyl-tyrosine)
to ascertain why this is so.
Commands:
-
select all (enter)
-
wireframe (enter)
-
select hydrophobic (enter)
-
colour white (enter)
-
select polar (enter)
-
colour red (enter)
-
select substrate (enter)
-
colour green (enter)
-
centre zn (enter)
-
Position the substrate molecule so that the phenolic hydroxyl group of
the substrate (in green) points away from you directly back into the screen.
You may wish to use the rotate command.
From the graphics window select sticks from the Display
menu. Since the substrate is the last selected item, it should now be displayed
as a stick structure.
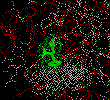
Let's now identify the amino acids that form a pocket to accomodate
the side chain of the carboxy-terminal amino acid (which in this case is
the phenolic group of tyrosine).
Commands :
-
define pocket 243, 250, 255, 203, 247 (enter)
-
select substrate or pocket (enter)
-
dots on (enter)
-
The dot outlines show the force field of Van der Waals contacts (Be patient
as it will take a few moments for the computer to calculate the field)
 Test
your knowledge!
Test
your knowledge!
-
Step Six - End of Part Two
-
Select the command window and enter the quit command


 .
When you see this icon,
single click on it to go to the appropriate
questions.
.
When you see this icon,
single click on it to go to the appropriate
questions.
![]()


 Test
your knowledge!.
Test
your knowledge!.


 Test
your knowledge!
Test
your knowledge!