








Main
Developmental biology aims to elucidate the molecular networks that regulate cell fate, organ formation and body response to endogenous signals or environmental biotic and abiotic factors. Arabidopsis thaliana has been established as the model plant to study the mechanisms underlying the growth or interactions of multicellular plant organisms with the environment. Genetics in combination with innovative molecular and cellular biology techniques provide fascinating insights towards the understanding of important aspects of plant development including cell fate determination, cell-to-cell communication, molecular signalling and transduction, response to environmental cues and plant plasticity. The major goal is to start thinking like a plant deciphering the origin and complexity of perplexed mechanisms leading to dead-ends or dynamic evolution in plant development.
Members
Lon Proteases
Quality control of proteins in eukaryotic organelles is predominantly maintained by members of the ATP-dependent proteases. Even though numerous biological analyses have shed light on the functional implications of such proteases, their involvement in developmental processes of multicellular organisms has not been determined. We recently identified two lon1 mutant alleles, both missing the carboxy terminal proteolytic domain, that show post-embryonic growth retardation resulting in delayed seedling establishment. These mutants provide novel insights into the developmental mechanisms of selective proteolysis in plant mitochondria. Further analysis aims to decipher the molecular recognition of protein substrates by Lon proteases and the events that control plant mitochondrial differentiation.
TRH1 Potassium Transporter
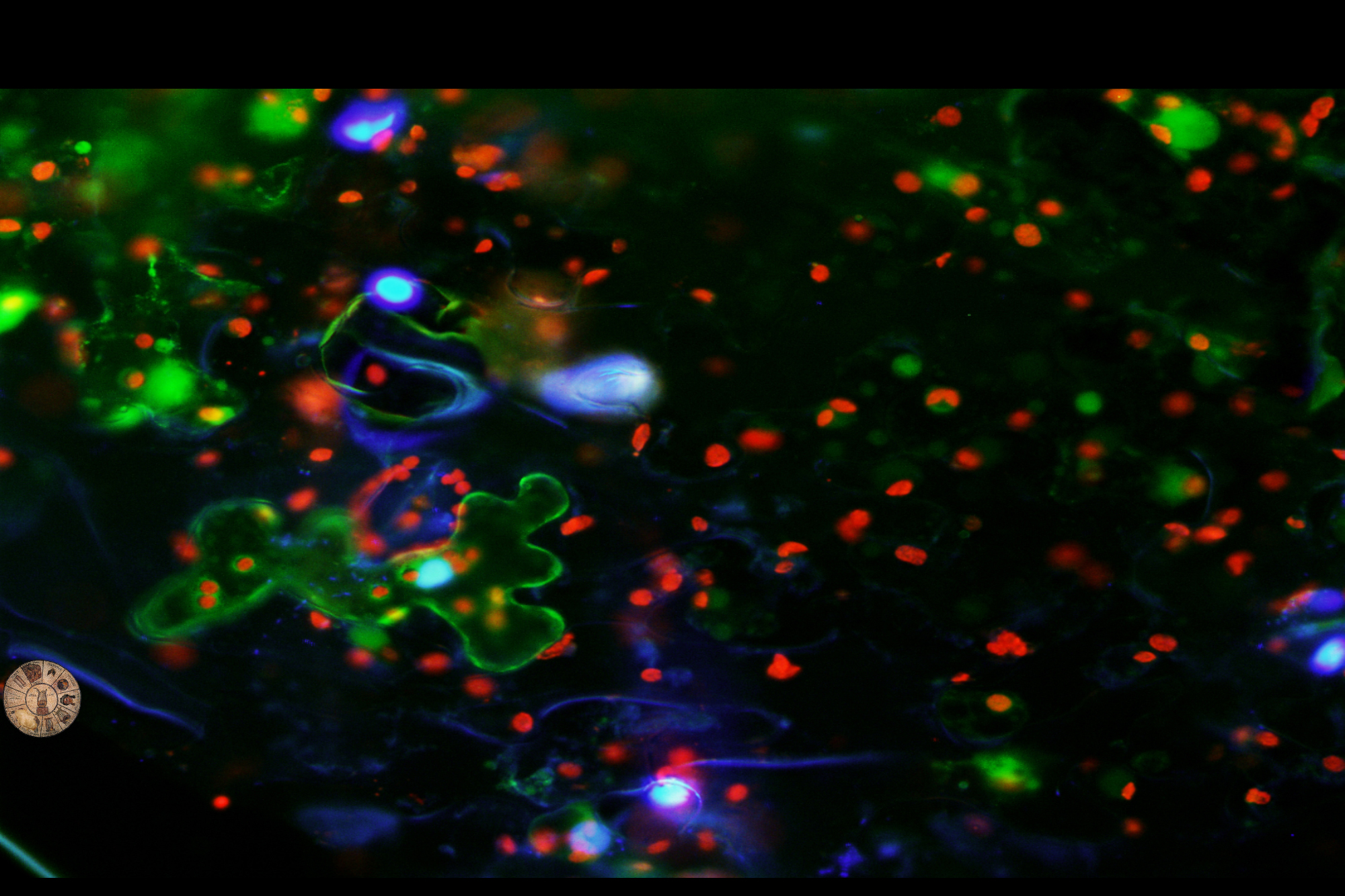
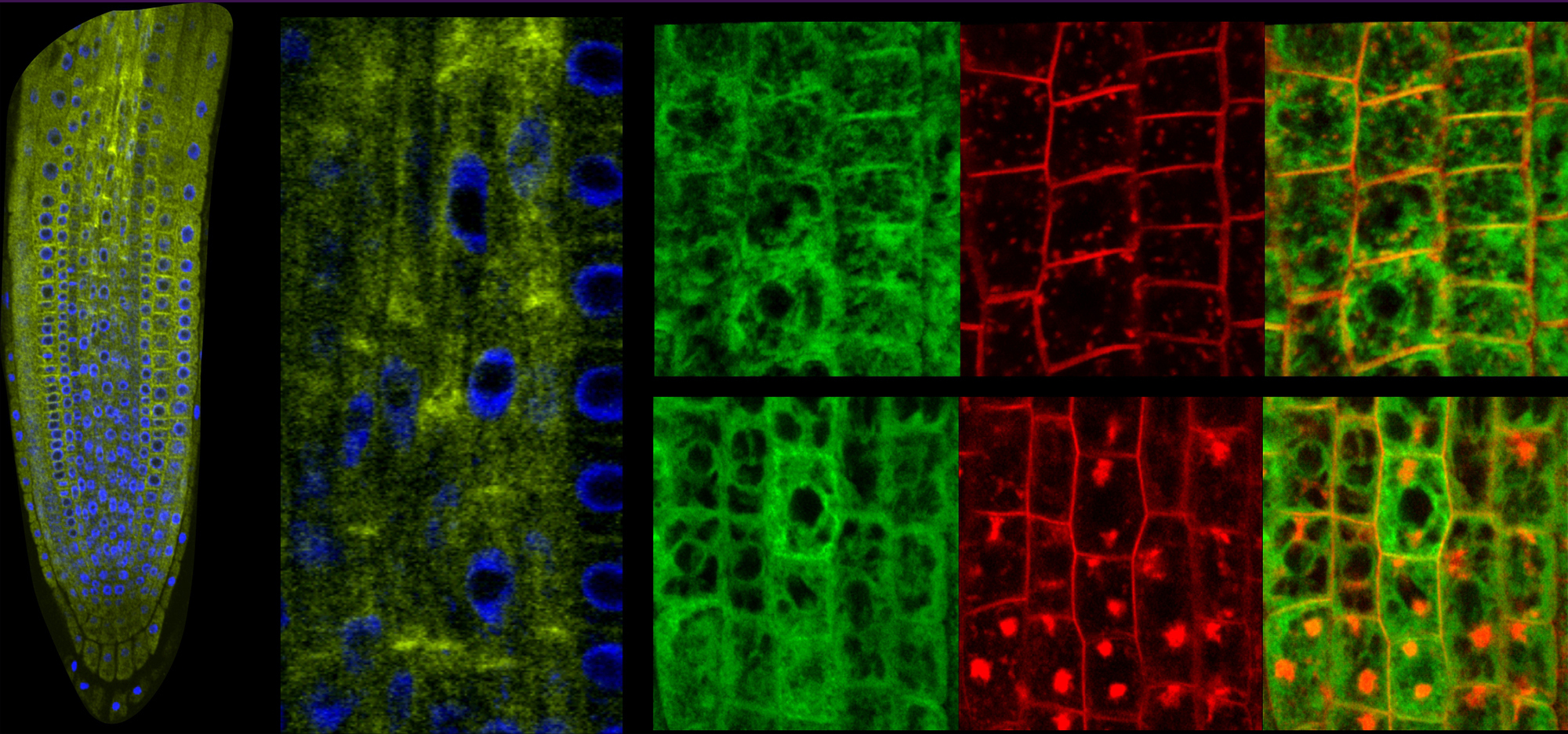
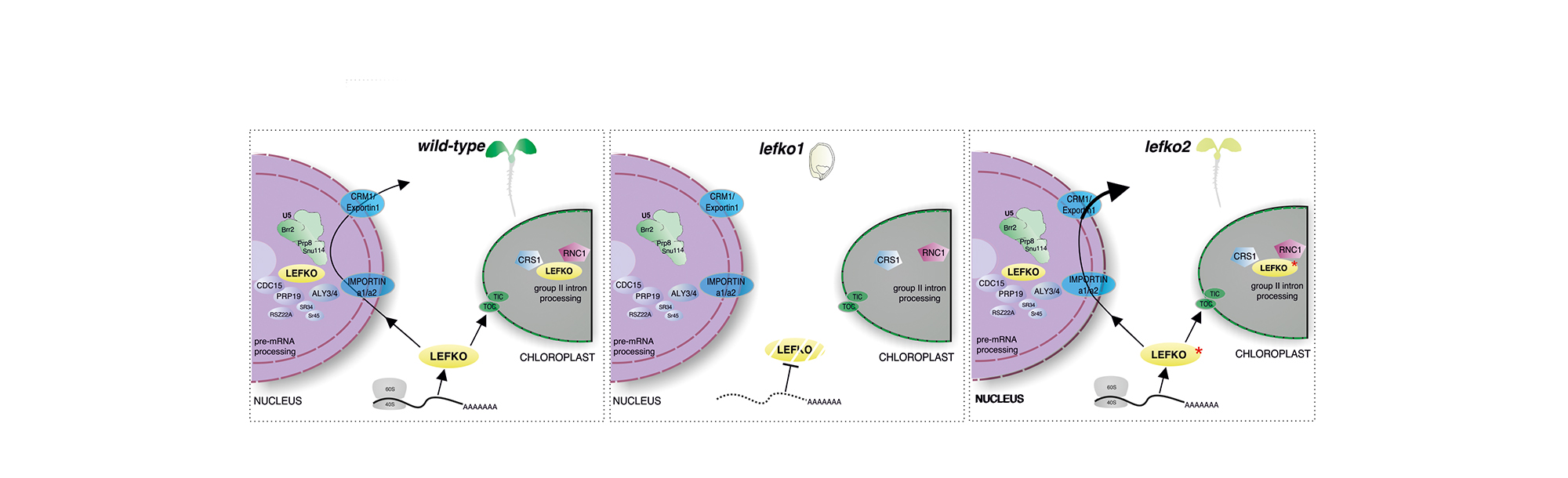
LEFKOTHEA RNA Binding Protein
Eukaryotic organisms accomplish the removal of non-coding sequences to produce mature mRNAs. Splicing mechanisms execute posttranscriptional programs in the nucleus and organelles and significantly affect development and homeostasis. Simultaneous deposition of the nucleus-encoded LEFKOTHEA protein in the nucleus and chloroplasts effectively commences the communication between the two organelles. LEFKOTHEA is a common component of the splicing activity of both chloroplast group II intron and nuclear spliceosome complexes, and converges the two splicing circuitries. This splicing factor substantially affects posttranscriptional outcomes to advance plant development. Transiently optimized LEFKOTHEA nuclear compartmentalization induces proper chloroplast biogenesis and lefko1 loss-of-function dismantles developmental progress by abruptly arresting embryogenesis. LEFKOTHEA provides a strong paradigm for the coevolution of splicing mechanisms of pre-mRNAs and group II introns and demonstrate that the convergence of the splicing networks in the nucleus and chloroplasts regulates plant growth.
Collaborations
Karin Ljung, Department of Forest Genetics and Plant Physiology, Swedish University of Agricultural Sciences, Sweden
Harvey Millar, Centre of Excellence in Plant Energy Biology and Centre for Comparative Analysis of Biomolecular Networks, University of Western Australia, Australia
Klaus Palme, Faculty of Biology, Albert-Ludwigs-University of Freiburg, Germany
Scott Poethig, Department of Biology, University of Pennsylvania, USA
Georgia Drakaki, Department of Plant Sciences, University of California, Davis, USA
George Panayotou, Biomedical Sciences Research Center, "Alexander Fleming",Greece
Emmanuel Panteris, Department of Biology, Aristotle University of Thessaloniki,Greece
Selected Publications
Koudounas K., Thomopoulou M., Michaelidis C., Zevgiti E., Papakostas G., Tserou P., Daras G. and Hatzopoulos P. (2017)The C-domain of oleuropein b-glucosidase assists in protein folding and sequesters the enzyme in nucleus. Plant Physiology
Daras G., Rigas S., Tsitsekian D., Zur H., Tuller T., Hatzopoulos P. (2014) Dual organellar targeting of Arabidopsis Lon1 by alternative transcription and translation initiation reveals the evolutionary status of twin presequences. Molecular Plant ssu030.
Rigas S., Daras G., Tsitsekian D., Alatzas A., Hatzopoulos P. (2014) Evolution and significance of the Lon gene family in Arabidopsis organelle biogenesis and energetic metabolism. Frontiers in Plant Science(5). Invited Review Article
Rigas S., Ditengou F., Ljung K., Daras G., Olaf T., Palme K., Hatzopoulos P. (2013) Root gravitropism and root-hair development constitute coupled developmental responses regulated by auxin homeostasis in the Arabidopsis root apex. New Phytologist 197(4):1130-41
Comment in: NEW PHYTOLOGIST 197(4):1027-8. -Pointing PINs in the right directions: a potassium transporter is required for the polar localization of auxin efflux carriers- Dolan, L.
Prassinos C., Rigas S., Kizis D., Vlahou A. and Hatzopoulos P. (2011) Subtle proteome differences identified between post-dormant vegetative and floral peach buds. Journal of Proteomics 74:607-619.
Rigas S., Daras G., Laxa M., Marathias N., Fasseas C., Sweetlove L.J. & Hatzopoulos P. (2009) The role of Lon1 protease in post-germinative growth and maintenance of mitochondrial function in Arabidopsis. New Phytologist
Comment in: NEW PHYTOLOGIST 181: 505-508. -Long bugs to short plants--the Lon protease in protein stability and thermotolerance- Taylor NL, Millar AH
Daras G., Rigas S., Penning B., Milioni D., McCann M.C., Carpita N.C., Fasseas C. and Hatzopoulos P. (2009) thanatos mutation in Arabidopsis cellulose synthase 3 (AtCesA3) has a semidominant-negative phenotype for cellulose synthesis and plant growth. New Phytologist184(1):114-26
Vicente-Agullo F., Rigas S., Desbrosses G., Dolan L., Hatzopoulos P., Grabov A. (2004) Potassium carrier TRH1 is required for auxin transport in Arabidopsis roots. Plant Journal 40: 523-535.
Rigas S., Debrosses G., Haralampidis K., Vicente-Agullo F., Feldmann K.A., Grabov A., Dolan A. and Hatzopoulos P. (2001) TRH1 encodes a potassium transporter required for tip growth in Arabidopsis root hairs. Plant Cell 13: 139-151.
Courses
(297) ΦΥΣΙΟΛΟΓΙΑ ΚΑΙ ΑΠΟΚΡΙΣΕΙΣ ΦΥΤΩΝ- PLANT PHYSIOLOGY -Lectures
(2790) ΒΙΟΛΟΓΙΑ ΚΥΤΤΑΡΟΥ-CELL BIOLOGY -Lectures -Lab Courses
(299) ΜΟΡΙΑΚΗ ΒΙΟΛΟΓΙΑ ΑΝΑΠΤΥΞΗΣ & ΚΑΤΑΠΟΝΗΣΕΩΝ ΤΩΝ ΦΥΤΩΝ -Lectures -Lab Courses
(3610) ΠΕΡΙΒΑΛΛΟΝΤΙΚΗ ΒΙΟΤΕΧΝΟΛΟΓΙΑ- ENVIRONMENTAL BIOTECHNOLOGY -ΕΡΓΑΣΙΕΣ -Lab Courses -Lectures
(205) ΒΙΟΤΕΧΝΟΛΟΓΙΑ ΦΥΤΩΝ-PLANT BIOTECHNOLOGY -Lab Courses
(234) ΕΙΔΙΚΑ ΘΕΜΑΤΑ ΒΙΟΤΕΧΝΟΛΟΓΙΑΣ ΦΥΤΩΝ-ΕΡΓΑΣΙΕΣ -Lab Courses -Lectures
(3606) ΜΟΡΙΑΚΗ ΒΙΟΛΟΓΙΑ & ΡΥΘΜΙΣΗ-ΕΡΓΑΣΙΕΣ -Lectures -Lab Courses
(3603) ΒΙΟΛΟΓΙΑ ΣΥΣΤΗΜΑΤΩΝ-ΕΡΓΑΣΙΕΣ -Lab Courses
Theses
Funding
ΕNIAIA ΔΡΑΣΗ ΚΡΑΤΙΚΩΝ ΕΝΙΣΧΥΣΕΩΝ ΕΤΑΚ ΕΡΕΥΝΩ-ΔΗΜΙΟΥΡΓΩ-ΚΑΙΝΟΤΟΜΩ
ΕΛΙΑΔΑ – Χαρακτηρισμός και αξιοποίηση γενετικού υλικού ελιάς με στόχο την προσαρμογή στην κλιματική αλλαγή και την παραγωγή υψηλής διατροφικής και οργανοληπτικής αξίας προϊόντων ελαιόλαδου – Τ2ΕΔΚ-01315
Services
Contact
Iera Odos 75, Athens, Greece, 118 55
